The Bagheri Lab integrates experimental data with computational strategies to elucidate fundamental properties governing intracellular dynamics and intercellular regulation. Our group is highly collaborative and integrates a diverse array of research interests. We take on grand challenges spanning complex dynamics of cell populations, to experimental design and tool development. A common thread that persists among our projects involves elucidating, predicting, and ultimately controlling biological response, particularly in context of disease. When the regulation, or control, of biological function fails, people can manifest a variety of illnesses including cancer and autoimmune disease.
Ongoing collaborative projects span three major thematic areas: emergent dynamics, focusing on how complex higher-level population dynamics arise from lower-level interactions within and among cells; machine learning, utilizing algorithms to learn patterns from data for prediction and analysis; and network theory, representing systems as graphs to study interactions and structure.
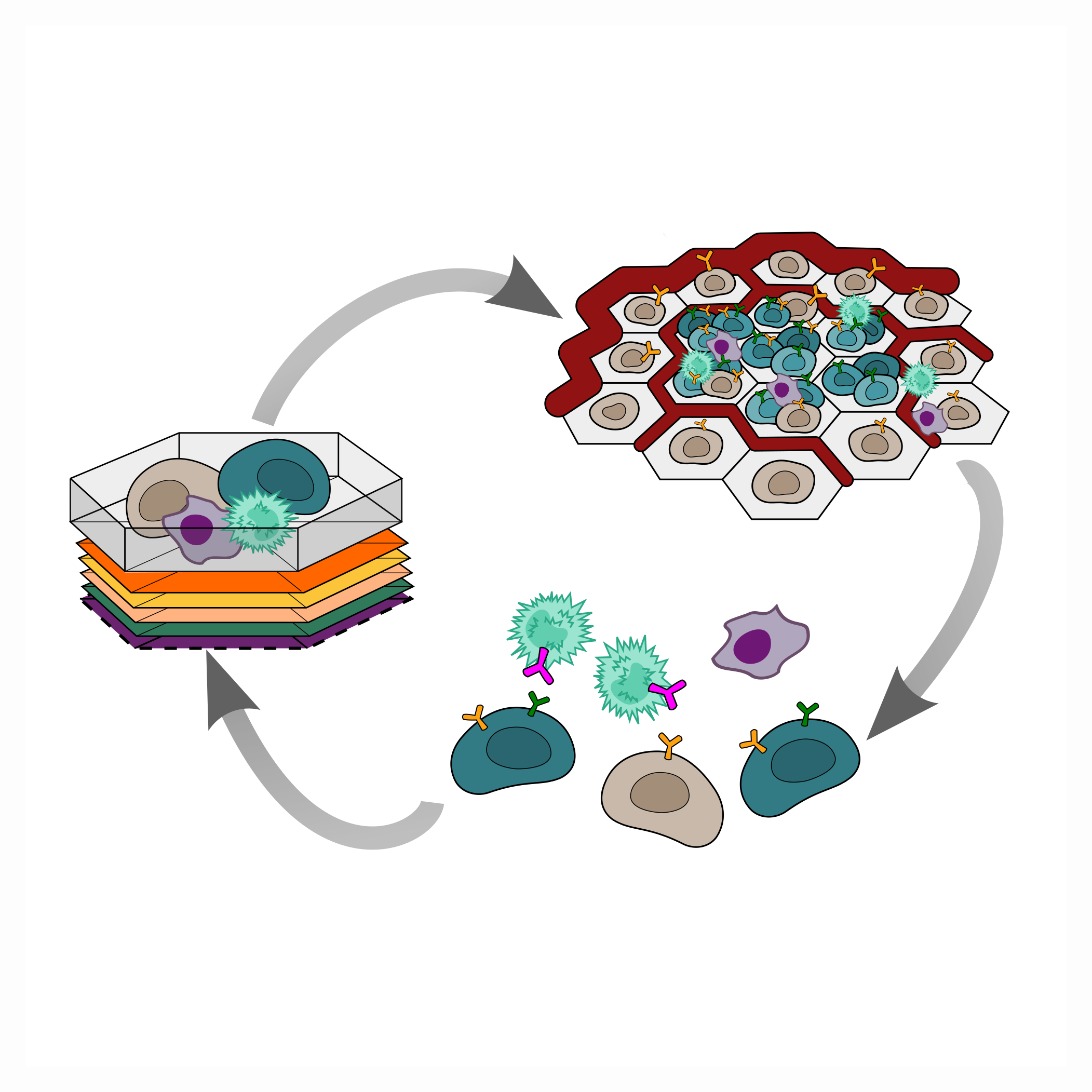
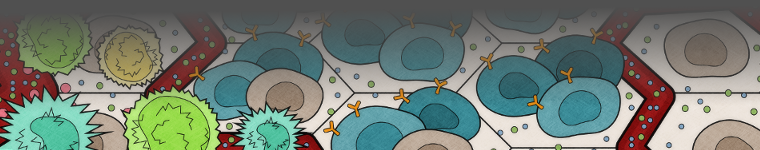
Combinatorial antigen cell therapy design
emergent dynamics
Chimeric Antigen Receptor (CAR) T cell therapy has demonstrated the power of designing endogenous T cells as a tool to target hematologic tumors. However, solid tumors introduce unique challenges due to diverse antigen presentation as well as the evolving solid tumor microenvironment (e.g. oxygen tension, immune privileged regions, angiogenesis). We build in silico models of cell-based therapies that combinatorially target multiple antigens in the context of a dynamic tumor microenvironment to broaden the viable targets for these therapeutic interventions.
Collaborators: M. B. Elowitz | M. G. Shapiro
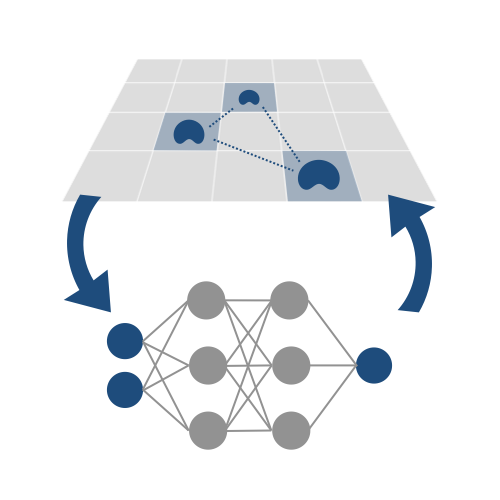
Representation learning and emulation of computationally expensive models
Jacob I. Evarts | Jason Y. Cain
machine learningemergent dynamics
Computational models aim to include enough biological detail to highlight important phenomena, but introducing too much detail can incur prohibitive computational costs. Running exhaustive simulations required for statistically rigorous analyses quickly becomes difficult or intractable. One possible alternative is to run exhaustive simulations on a separate model, trained to learn key dynamics of the original simultion from low dimensional representations. We identify the appropriate learning goals to replicate the emergent function of interest as a key step to stengthening insights we can extract from agent-based models.
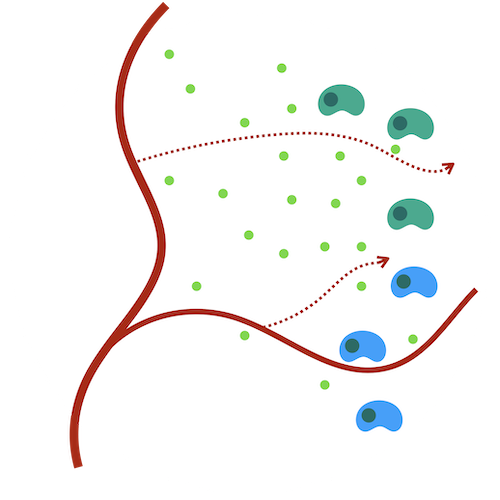
Hypoxia-angiogenesis feedback loop and tumor development
emergent dynamicsnetwork theory
Hypoxia, a hallmark of most solid tumors, is highly associated with poor patient outcomes. Hypoxic conditions lead to cells secreting angiogesis factors, providing cytokines to support blood vessel development and nutrient distribution. This relationship results in hypoxia being both a driver and a consequence of cancer progression. An accurate, robust model of the multiscale spatio-temporal relationship between hypoxia and angiogenesis is critical to guiding interventions and interrogating the subsequent emergent dynamics.
Lateral root development
emergent dynamics
Root system architecture is a major determinant of plant fitness. The ability to precisely control root system architecture would enable farmers to optimize plant fitness for field conditions, maximizing production efficiency. We aim to interrogate mechanisms underlying cellular decision making during lateral root development in dicot plants using an agent-based model. We will calibrate this model to ensure it accurately captures relevant developmental dynamics and then use it to identify control inputs and perturbations that allow for the precise control of lateral root formation.
Collaborators: J. Nemhauser

Spatial structure and eco-evolutionary stability of microbial consortia
emergent dynamics
Most microbes live in cooperative consortia, where species specialize in one task and depend upon their neighbors to perform tasks they cannot. A microbe is only successful if it is surrounded by supportive neighbors, and yet the best strategy for any microbe is to *not* provide support to others. These two factors—ecological context and evolved traits—determine the eco-evolutionary stability of a consortia. Here I use in silico modeling to explore how the degree of spatial structure in a community affects its ability to persist ecologically and avoid collapse evolutionarily due to the invasion of selfish variants.
Collaborators: B. Kerr
Multiscale, multiclass models of cell populations
emergent dynamics
To effectively understand and predict cell population responses to intrinsic perturbations and extrinsic intervention, the integration of intracellular signaling with intercellular dynamics is necessary. Through agent-based models, we simulate and predict the behavior of heterogeneous cell populations. Our in silico framework (developed by Jessica S. Yu) effectively elucidates how changes at the subcellular level emerge into varied population level responses.

Asymmetrical cell division in Drosophila neurogenesis
emergent dynamicsnetwork theory
Neurogenesis is the process by which neurons are generated from neural stem cells. In Drosophila, neurogenesis requires a series of asymmetrical cell divisions that yield daughter cells with distinct sizes and identities. The mechanisms determining the intracellular locations of these cell divisions and how these distinct identities are established are not yet fully understood. We are developing an agent-based model to explore the dynamics of cell division and differentiation during Drosophila neurogenesis, with the goal of identifying the key factors that regulate where cell division occurs and how daughter cell identities are determined.
Collaborators: C. Cabernard

Propagation of variance in heterogeneous spatiotemporal models
emergent dynamics
Intrinsic and extrinsic variance are known to be inherent to biological systems and play a critical role in processes underlying tumor growth/evolution, tissue repair, and cell migration. Computational models have effectively replicated dynamics that emerge from stochastic rules/equations, but the impact of these sources of numerical variance on simulated dynamics across temporal and spatial scales has yet to be uncovered. This project unpacks the propagation of variance and quantifies how cell-level sources of intrinsic and extrinsic variance drive population level outcomes in high-resolution spatiotemporal agent-based models.